Evangelos Kolettas
Affiliated Researcher/ BRI Group Leader, Biomedical Research Institute, FORTH, Professor of Molecular Cell Biology, and Head of the Department of Biology, School of Medicine, University of Ioannina, Ioannina
Molecular Cancer Biology and Senescence (MCBS) Research Group
My laboratory has two research directions: (a) Cell signalling and regulatory networks in DNA damage and inflammation involved in senescence, and cancer, with a focus on IKK/NF-κB transcription factor (TFs) signalling pathways, and (b) Functional domain-specific CRISPR/Cas9 screens to identify novel regulators of cell growth, drug tolerance and resistance of cancer cells.
(Α) Cell Signalling and Regulatory networks in DNA damage and inflammation impacting senescence and cancer
Members of the IKK/NF-κB TF family are pivotal regulators of immune, pro-inflammatory and stress-like responses in health and disease and have been involved in DNA damage responses and inflammation impacting senescence and cancer, and also chronic inflammatory diseases.
IKK/NF-κB TFs are involved in both extracellular (e.g., pro-inflammatory cytokines), and in intracellular signalling (e.g., ROS, DNA damage) leading to the activation or suppression of target gene expression. They are activated mainly by two signalling pathways: a canonical (IKKβ-RelA(p65)/p50) and a noncanonical (IKKα-RelB/p52) signalling pathway, in response to both extracellular and intracellular stimuli. In addition, the upstream NF-κB activating Ser/Thr kinases IKKα and IKKβ are also involved in the regulation of fundamental cellular processes, with diverse physiological effects in different cells, independently of the activation of the NF-κB TFs.
IKK/NF-κB signaling components are aberrantly expressed and/or activated in pulmonary diseases and in non-small cell lung cancer (NSCLC), with an unfavourable prognosis for patient survival. We have investigated the mechanisms of action of IKKβ-RelA(p65) and IKKα-p52 signalling in NSCLC in vitro and in vivo, by employing biochemical, cell and molecular biology techniques, bio-imaging and -omics technologies in conjunction with bioinformatics. We have generated novel lung-specific bi-transgenic Sftpc-CreERT2:ROSA-fLacz (IKKαf/f and/or IKKβf/f) mouse strains expressing a tamoxifen (Tmx)-inducible Cre recombinase (Sftpc-CreERT2) under the transcriptional control of the lung epithelial cell-specific surfactant protein C (SPC) promoter in alveolar type II (AT-II) lung epithelial cells (Chavdoula et al 2019 Life Sci Alliance). We have also generated inducible IKKα or IKKβ knock-down human NSCLC lines using lentiviral vectors, and IKKα or IKKβ knock-out human NSCLC lines using CRISPR/Cas9 lentiviral vectors. These novel in vivo and in vitro research tools are being used to study the impact of each IKK on DNA damage and inflammation on NSCLC and senescence.
We showed that canonical IKKβ-RelA(p65) signalling promotes tumour growth by downregulating the membrane-bound metastasis suppressor CD82/TSPAN27, affecting epithelial-to-mesenchymal cell transition (EMT) via integrin-mediated signalling (Roupakia et al 2021 Cancers) (Fig. 1). In addition, RelA(p65) and E2F1 cistromes have limited overlap and bind active chromatin even prior to immunogenic stimulation (Foutadakis et al 2022 Cancers).
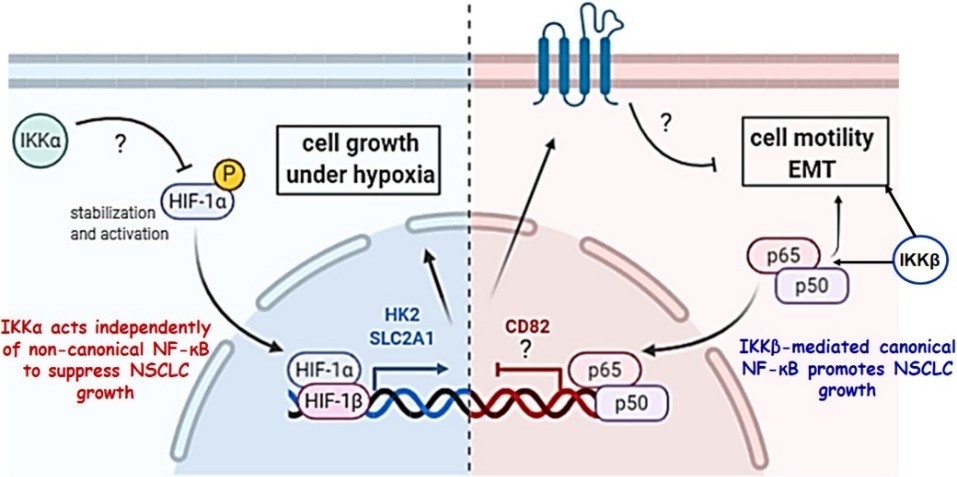
Fig. 1. Mechanisms of action of IKKα-p52 and IKKβ-p65 in NSCLC development and progression. In contrast to NF-κB p52 subunit, IKKα acts independently of non-canonical NF-κB to suppress NSCLC growth. IKKβ-mediated canonical NF-κB promotes NSCLC growth.
We also showed that, in contrast to canonical NF-κB signalling pathway and the noncanonical NF-κB p52 subunit acting as tumour promoters, IKKα acts as a tumour suppressor (TS) by regulating HIF and hypoxia-mediated processes, including carbohydrate metabolism and NRF2 oxidative stress pathways required for the in vivo human cancer cell growth as tumour xenografts and in mouse NSCLC growth in response to urethane, a tobacco constituent (Chavdoula et al (2019) Life Sci Alliance) (Fig. 1).
Defining mechanisms of IKKα’s tumour suppressor function in human NSCLC: We continue working on the mechanism(s) of the pleiotropic action of IKKα, a nucleocytoplasmic protein, as TS in human NSCLC. We characterised the IKKα interactome and performed deep transcriptome analysisn IKKαKO vs IKKαWT control human NSCLC cells generated by CRISPR/Cas9 lentivectors. We will use human NSCLC lines, patient derived xenografts and our novel lung-specific bi-transgenic, Spc-CreERT2:IKKαf/f mouse models to induce NSCLC by urethane, and employ a multi-omics integrative approach including state-of-the-art high throughput -omics technologies to address important questions from different angles, aiming to define the novel function of IKKα in controlling gene expression, accounting for IKKα’s tumour suppressor activity in NSCLC.
Future project:
- Mutant oncogene-induced cellular and metabolic reprogramming affecting chromatin remodelling in NSCLC
- Role of the regulatory NF-κB-miRNA network in NSCLC
- Crosstalk between DNA damage responses and mitotic spindle assembly checkpoint.
- Stress-induced changes in splicing and translation in immunity and cancer
- Mechanisms of action of IKKα in senescence
(Β) Functional domain-specific CRISPR/Cas9 screens to identify novel regulators of cell growth and behaviour.
A second research direction involves functional domain-specific CRISPR/Cas9 screens such as kinase, TFs and epigenetic regulators to identify novel regulators of cell growth and behaviour, including novel regulators of DNA damage and inflammation involved in senescence and cancer.








